Prediction of solvation free energies and solvent/water partition coefficients using molecular dynamics simulations
We have developed, in collaboration with Oliver Beckstein (Arizona State University, USA), three tools to facilitate the computation of solvation free energies (ΔGsolv) and of solvent/water partition coefficients (logKSW) using molecular dynamics simulations :
- MDPOW – computation of solvation free energies (ΔGsolv) and of solvent/water partition coefficients (logKSW) ;
- MOL2FF – automatic parameterization of ligands using OPLS-AA force field ;
- Ligandbook – online repository for topologies and force field parameters of ligands.
More generally, these tools are essential for facilitating the molecular dynamics simulations involving protein-ligand complexes.

Representation of the three tools developed in the team (MDPOW, MOL2FF and Ligandbook) in connection with molecular dynamics simulations.
Participation in international SAMPL prediction competitions
Using the MDPOW software and the OPLS-AA force field parameters obtained using the MOL2FF, we have participated in the SAMPL3 (2011), SAMPL4 (2013) and SAMPL5 (2015) international prediction competitions, generally obtaining very good results.
SAMPL3 (2011)
Using this protocol, in 2011 we took part in the SAMPL3 international prediction competition, which asked us to predict ΔGhyd values for 36 molecules organised in three series, derived from ethane, biphenyl and dibenzo-p-dioxin, bearing a variable number of chlorine atoms. The decisive factor that set us apart from the other participants was not only to use the available OPLS-AA parameters, which were not very suitable for molecules carrying a large number of chlorine substituents, but also to develop new parameters, with charges obtained by quantum calculations using two different methods. We were thus able to develop a new set of transferable parameters for molecules with more than three chlorinated substituents on an aromatic ring.
Under these conditions we were able to predict ΔGhyd values for these difficult compounds with a root mean square error (RMSE) of 1.01 kcal/mol (compare with an RMSE of 4.27 kcal/mol obtained with the original OPLS-AA parameters) and one of our solutions was ranked first in the ‘Hydration free energy prediction’ section.
SAMPL4 (2013)
A similar protocol was used in 2013 to predict the hydration free energy of 52 compounds with very diverse structures, as part of the SAMPL4 competition. We were able to parameterise two new chemical functions, N-alkyl-imidazole and nitrate, and in some cases highlight problems with the reproducibility of the Lennard-Jones free energy term, which are mainly linked to the conformational flexibility of the molecule and the starting conformation. Overall, our prediction was made with a very acceptable accuracy (RMSE compared with experimental values) of 1.68 kcal/mol.
SAMPL5 (2015)
As part of the SAMPL5 competition we predicted the cyclohexane/water distribution coefficients logKCW for 53 organic molecules, using MDPOW software and OPLS-AA force field parameters generated by MOL2FF. This work also enabled us to validate the OPLS-AA force field parameters for cyclohexane, a sine qua non for making good predictions. The standard error (RMSE) of our predictions was almost 4 log units, while the mean error (ME) was -3 compared with the experimental values. These values, which are similar to those obtained by the other participants, are not as good as those expected by the organisers and highlight the difficulties in predicting the solvent/water distribution coefficients. The average error we obtained, which is very far from the expected zero value, suggests the presence of a systematic error in our predictions, but only the experimental values of logKCW do not allow us to identify the precise source of this error. Preliminary results we have recently obtained suggest that this error comes from the free energy of hydration.
Generally speaking, these participations in the SAMPL competitions have enabled us to parameterise several new chemical functions and to highlight a number of problems linked to the protocol and the use of the OPLS-AA force field, in particular a systematic error in the calculation of the hydration free energy, which is currently being investigated.
Articles :
- Kenney, I. M. ; Beckstein, O. ; Iorga, B. I., Prediction of cyclohexane-water distribution coefficients for the SAMPL5 data set using molecular dynamics simulations with the OPLS-AA force field. J. Comput. Aided Mol. Des. 2016, 30, 1045-1058 [Online version].
- Beckstein, O. ; Fourrier, A. ; Iorga, B. I., Prediction of hydration free energies for the SAMPL4 diverse set of compounds using molecular dynamics simulations with the OPLS-AA force field. J. Comput. Aided Mol. Des. 2014, 28, 265-276 [Online version].
- Beckstein, O. ; Iorga, B. I., Prediction of hydration free energies for aliphatic and aromatic chloro derivatives using molecular dynamics simulations with the OPLS-AA force field. J. Comput. Aided Mol. Des. 2012, 26, 635-645 [Online version].
Conference proceedings :
- Beckstein, O. ; Iorga, B. I. Prediction of hydration free energies for aliphatic and aromatic chloro derivatives using molecular dynamics simulations with the OPLS-AA force field, 243rd ACS National Meeting & Exposition Abstracts of Papers, San Diego, CA, United States, March 25-29 ; American Chemical Society : San Diego, CA, United States, 2012 ; pp COMP-76.
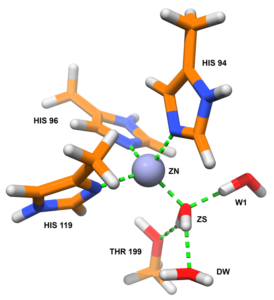
Parametrisation of the active sites of metallo-enzymes in the OPLS-AA force field
Using quantum calculations and a protocol developed in the team, we parameterised the residues making up the active site of carbonic anhydrase. The main difficulties were the presence of the zinc ion and the two protonation states of the ZS water molecule. These parameters were then validated by molecular dynamics simulations.
In a similar way, we are currently in the process of parameterising the active sites of the 3 sub-classes (B1, B2 and B3) of metallo-β-lactamases that contain one (B2) or two (B1 and B3) zinc ions.
Articles :
- Bernadat, G. ; Supuran, C. T. ; Iorga, B. I., Carbonic anhydrase binding site parameterization in OPLS-AA force field. Bioorg. Med. Chem. 2013, 21, 1427-1430 [Online version].
Development of an efficient docking protocol for virtual screening
SAMPL3 (2011)
As part of our participation in the 2011 SAMPL3 prediction competition in the ‘Virtual fragment screening’ section, from a library of 500 fragment ligands we were asked to rank these ligands according to their affinity for bovine trypsin. The key step that set us apart from the other participants was the prior optimisation of the software and docking parameters in relation to the system to be studied. We extracted all known trypsin inhibitors from the literature, along with their biological activity, and selected those of the fragment type. We also analysed all available X-ray structures of trypsin complexed with fragment ligands. Docking these ligands onto the bovine trypsin structure with the best resolution, using two docking software packages (Gold and Glide) and several scoring functions, enabled selection of the software package and scoring function that led to the best correlation with the biological data. Generally speaking, Gold gave very good results, while Glide’s results were rather disappointing. Using this optimised protocol, we docked all 500 fragments, analysed the interactions within the site and generated AUC ROC curves. Another aspect that probably differentiated us from the other participants was the use of typical conformational search parameters for docking (100%) and not those typical for virtual screening (30%). Even if the calculation time was longer, the better quality of the results obtained largely justified this choice.
Using the docking protocol optimised for virtual fragment screening as described above, we submitted three solutions which were ranked in the top three places in the “Virtual fragment screening” section. One of our images was selected to illustrate the cover of the special issue of J. Comput. Aided Mol. Des. dedicated to this competition.
SAMPL4 (2013)
A similar protocol was used in 2013 for the virtual screening section of the SAMPL4 competition. The challenge was to predict the interaction of 321 molecules with 3 different HIV integrase sites. The main difficulties encountered by all the participants were linked to the high structural similarity between the active and inactive compounds, as well as the presence of 3 sites. One of our predictions came second in the competition.
CSAR (2014)
As part of our participation in the CSAR 2014 competition, using a protocol developed in the team we correctly predicted by docking all 22 protein-ligand complexes requested in Phase 1. The key points that enabled us to achieve this very good result were: i) preliminary analysis of the docking software and score functions available in order to choose the combination that is best suited to the target protein and ii) improved parameters for conformational search during docking, at the cost of a much longer computation time, in order to better explore the conformational space of large and flexible ligands. In Phase 2, we proposed docking poses with an average RMSD of 1-2 Å, depending on the characteristics of each series. However, ranking the poses proved more problematic, highlighting the limitations of the score functions currently available. An image from this work was selected for the cover of the special issue of J. Chem. Inf. Model. dedicated to the CSAR competition. Our publication was the most widely read open access ACS AuthorChoice article in the journal J. Chem. Inf. Model. in 2016.
D3R Grand Challenge (2015)
As part of our participation in the D3R Grand Challenge 2015 competition, we were asked to predict the relative affinity of 180 ligands of Heat Shock Protein 90 (HSP90) and the interaction mode for 6 of them, as well as the interaction mode for 30 ligands of Mitogen-Activated Protein Kinase Kinase Kinase 4 (MAP4K4) and predict the relative affinity for 18 of them. The HSP90 ligands belonged to three families (benzimidazolones, aminopyrimidines and benzophenone derivatives), while the MAP4K4 ligands were very diverse.
A thorough analysis of the available experimental data (structural and biochemical) showed that Gold with the GoldScore function gave the best results with these two proteins, and that residues Lys58 (HSP90) and Lys54 (MAP4K4) must be flexible during docking. In the case of HSP90, docking was performed on 11 structures representative of the conformations of the 191 crystallographic structures available in the Protein Data Bank (PDB). In this way, we were able to better explore the conformational space of this protein, which is very flexible in the region of residues 99-129.
The prediction of the interaction mode of the first four ligands with HSP90 was excellent, with the exception of the pyridylsulphonyl group in HSP044, while for the last two (HSP175 and HSP179) we predicted the orientation in the active site well overall, but with slightly different interactions. These results correspond to an average RMSD of 1.48 Å for the conformations ranked first, and an average RMSD of 1.20 Å for the poses with the best RMSD. For MAP4K4, we correctly predicted 11 ligands that exhibited a ‘classical’ interaction mode and incorrectly predicted 17 ligands that exhibited a novel interaction mode and 2 ligands that interacted with the active site through water molecules.
As with previous competitions, predicting the relative affinities of ligands was more difficult for most participants. However, our submission for MAP4K4 was ranked 2nd out of 46 submissions.
This work has highlighted the need to identify the docking tools and parameters that are best suited to each target, as well as the active site residues that need to be flexible during the docking process. Further processing of the docking poses, in particular by free energy calculations, seems necessary to improve the prediction of the relative affinity of the ligands, in particular by taking into account the influence of water molecules and the overall flexibility of the protein, and our future efforts will be concentrated in this direction.
Articles :
- Selwa, E. ; Martiny, V. Y. ; Iorga, B. I., Molecular docking performance evaluated on the D3R Grand Challenge 2015 drug-like ligand datasets. J. Comput. Aided Mol. Des. 2016, 30, 829-839 [Online version].
- Martiny, V. Y. ; Martz, F. ; Selwa, E. ; Iorga, B. I., Blind pose prediction, scoring, and affinity ranking of the CSAR 2014 dataset. J. Chem. Inf. Model. 2016, 56, 996-1003 [<ahref=”http://dx.doi.org/10.1021/acs.jcim.5b00337″>Online version] (Open Access).
- Colas, C. ; Iorga, B. I., Virtual screening of the SAMPL4 blinded HIV integrase inhibitors dataset. J. Comput. Aided Mol. Des. 2014, 28, 455-462 [<ahref=”http://dx.doi.org/10.1007/s10822-014-9707-5″>Online version].
- Surpateanu, G. ; Iorga, B. I., Evaluation of docking performance in a blinded virtual screening of fragment-like trypsin inhibitors. J. Comput. Aided Mol. Des. 2012, 26, 595-601 [<ahref=”http://dx.doi.org/10.1007/s10822-011-9526-x”>Online version].